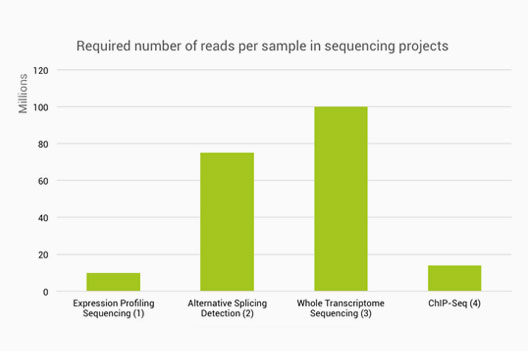
Frontiers | Optimization of an RNA-Seq Differential Gene Expression Analysis Depending on Biological Replicate Number and Library Size
Evaluation of critical data processing steps for reliable prediction of gene co-expression from large collections of RNA-seq data | PLOS ONE
RnaSeqSampleSize: real data based sample size estimation for RNA sequencing | BMC Bioinformatics | Full Text
Transcriptome diversity is a systematic source of variation in RNA-sequencing data | PLOS Computational Biology
BioMedInformatics | Free Full-Text | Adjusted Sample Size Calculation for RNA-seq Data in the Presence of Confounding Covariates
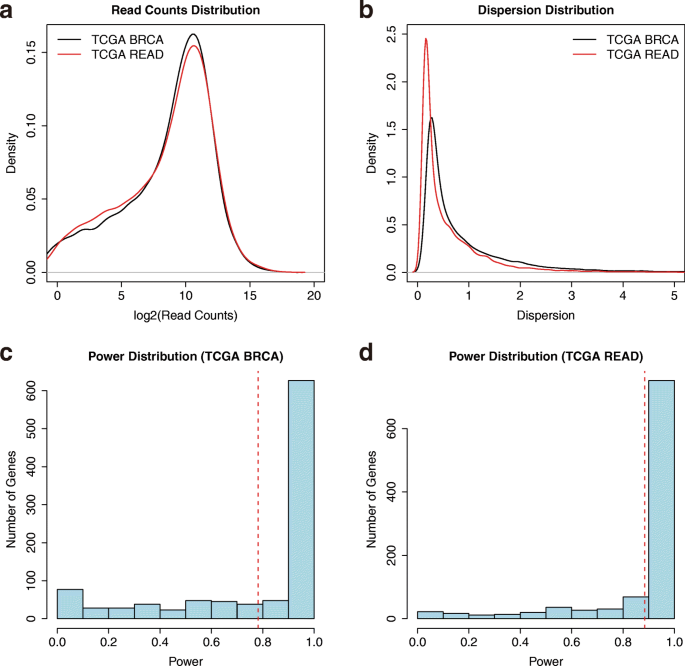
RnaSeqSampleSize: real data based sample size estimation for RNA sequencing | BMC Bioinformatics | Full Text
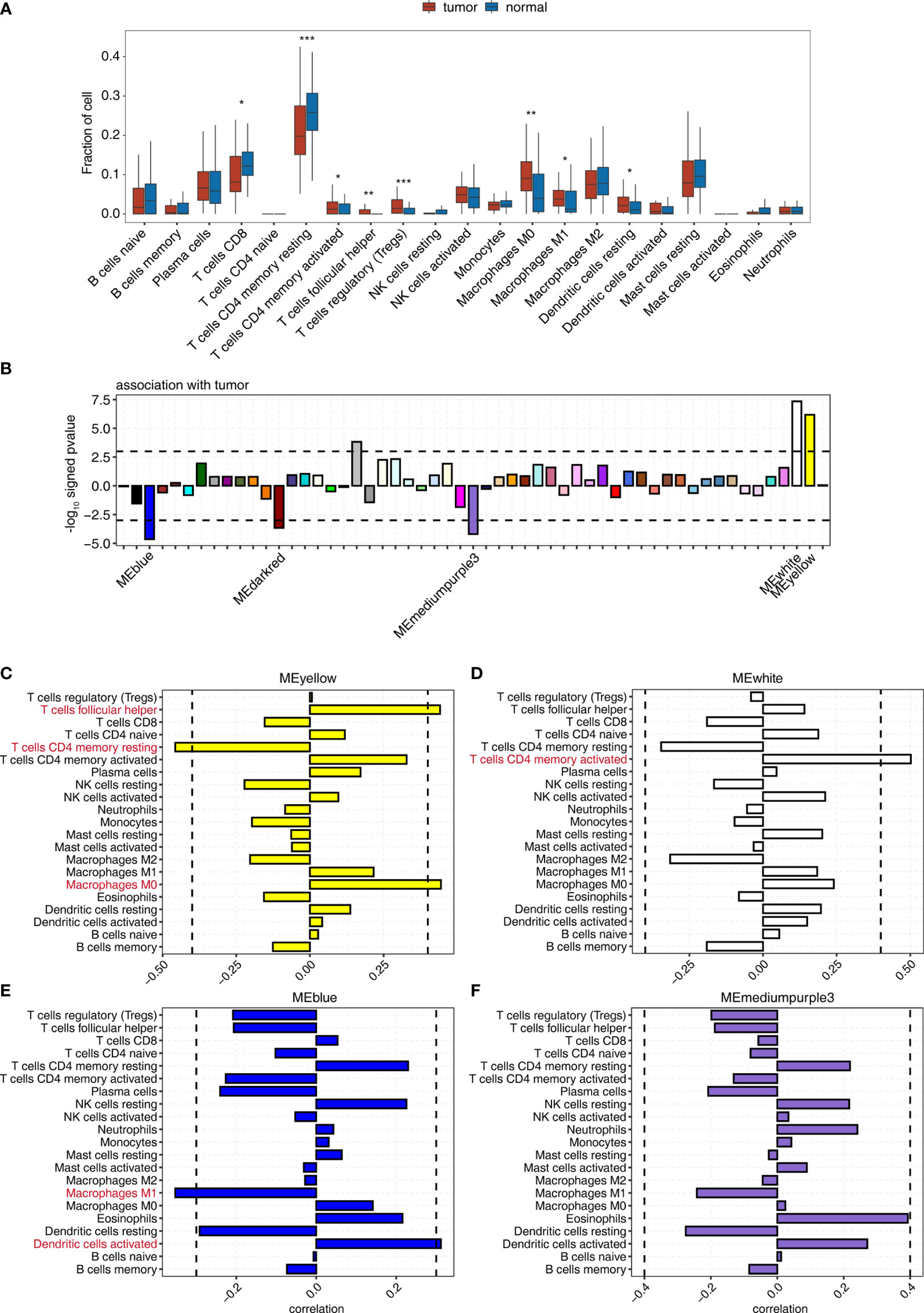
Frontiers | Analysis of Bulk RNA Sequencing Data Reveals Novel Transcription Factors Associated With Immune Infiltration Among Multiple Cancers

Longitudinal RNA-Seq Analysis of the Repeatability of Gene Expression and Splicing in Human Platelets Identifies a Platelet SELP Splice QTL | Circulation Research

NASA GeneLab RNA-seq consensus pipeline: Standardized processing of short-read RNA-seq data - ScienceDirect
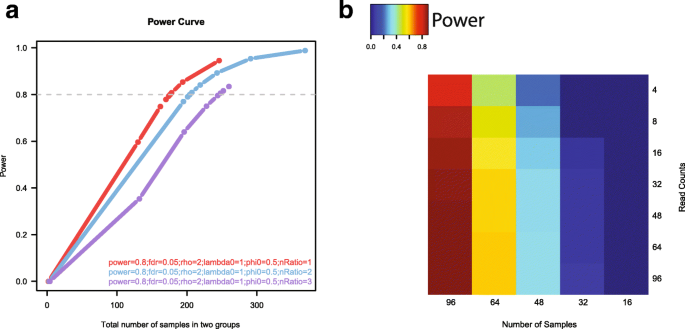
RnaSeqSampleSize: real data based sample size estimation for RNA sequencing | BMC Bioinformatics | Full Text
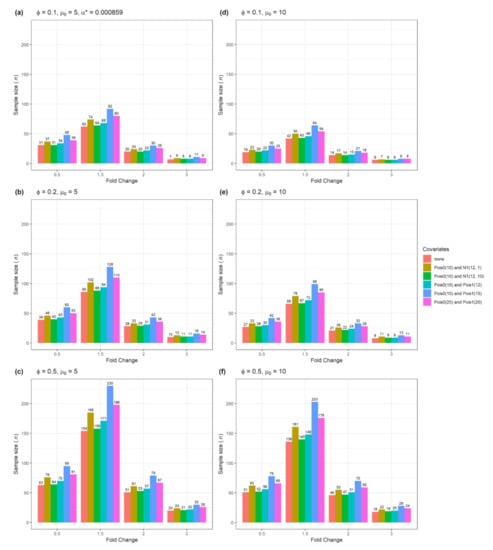
BioMedInformatics | Free Full-Text | Adjusted Sample Size Calculation for RNA-seq Data in the Presence of Confounding Covariates